The QBRC provides data analysis and data mining services for -omic and high dimensional data sets using bioinformatics, systems biology, and machine learning techniques. The QBRC staff will assist investigators on planning, analyzing, and interpreting the resulting data and provide publication ready materials. We are particularly equipped to analyze “Big Data” and to implement complex algorithms using secure Cloud Computing infrastructure. The QBRC also provides research software engineering (RSE) services.
We provide the following four types of services:
Data Mining
Pattern Classification
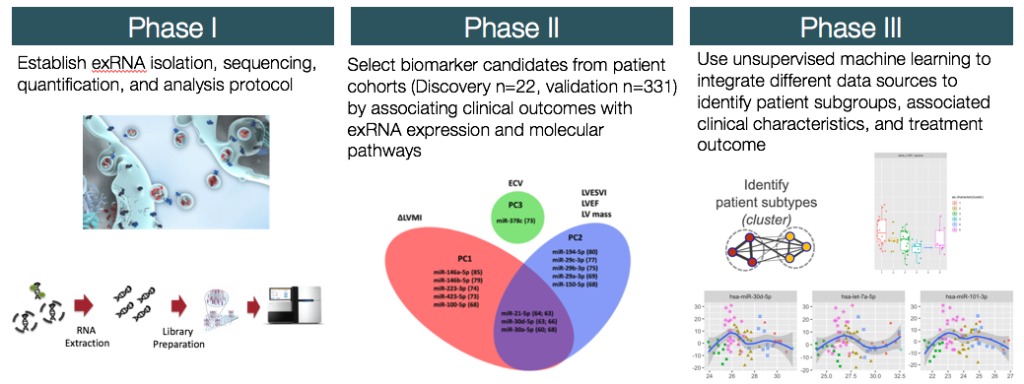
We offer highly customized data mining services using machine learning tools such as regression models, unsupervised clustering methods, and supervised learning methods. These type of projects include but not limited to:
- Population subtype discovery
- Biomarker discovery
- Predictive model construction
Please refer to our publications or contact us for more information.
Multi-Data Type Integration
Technology advancements have made it cost-effective to collect diverse types of high dimensional data. We offer network-based computational methods to combine multiple data types, such as clinical, questionnaires, image, and genomic data, for a set of samples to perform pattern recognition and model interactions.
- Identify sample similarity and community from multi-type data measurements
- Assess level of interaction between driver and respondents (i.e. gene expression driven by transcription factor binding)
- Isolate single sample contribution to network
Image Analysis using Deep Learning
Complex deep convolutional neural networks (CNNs), which are able “automatically” to capture predictive features in image data once appropriately trained, have garnered significant attention for their potential utility in biomedical applications with its accuracy. We provide deep learning neural network model construction service for quantitative image analysis. We currently support high-throughput microscopy assays using state-of-the-art convolution neural networks (CNNs) and related techniques for model construction.
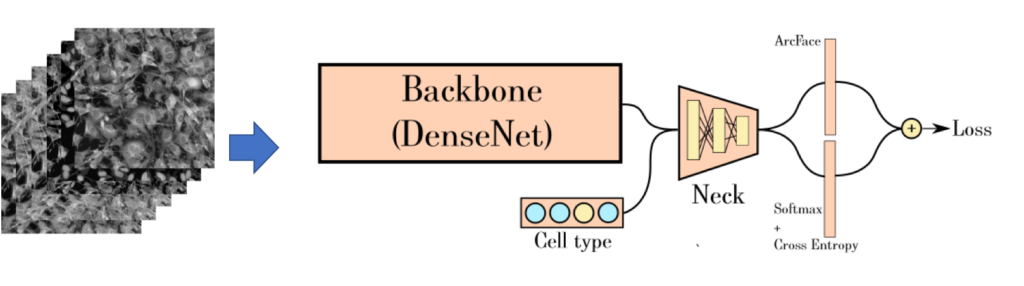
Please refer to our case study or contact us for more information.
Genomic Data Analysis
We offers a wide range of easy-to-use standard Next Generation Sequence data analysis pipelines that can rapidly process large number of samples (>100) from raw files, analyses include:
- Germline/Somatic Mutation Analysis
- Single Cell RNA-Seq Analysis
- Bulk RNA-Seq
- Small RNA-Seq
- ChIP-Seq
Please refer to our CNAP platform or contact us for more information.
Customized Analysis Pipeline Construction
We offers customized analysis pipeline construction on our cloud computing infrastructure that offers fast deployment, easy dissemination, and the ability to process large data volume in a secure and highly reproducible environment.
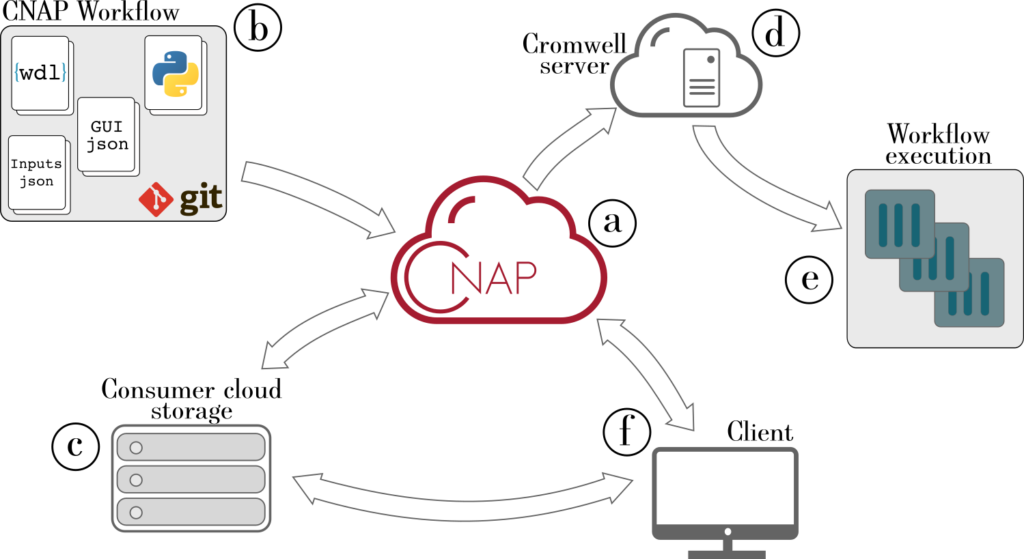
Research Software Engineering
Do you need help with software or cloud engineering tasks that distract you from doing science? We can accelerate your research by developing usable, robust, and maintainable scientific software, and more!
We can assist with:
-
- Software design, implementation, optimization, refactoring, and maintenance
- Every phase of the research software engineering life cycle from pre-publication to post-publication
- Cloud computing
- From budgeting for grant applications to implementing, securing, and maintaining infrastructure as code
- Data management
- Storage and sharing of large ‘omics data sets
- Data visualization
- interactive dashboards using R Shiny and other tools
- Software package management
- Containers or Conda packages for your software tools to facilitate distribution
- Many other areas of scientific computing
- HPC, web development, etc.
- Software design, implementation, optimization, refactoring, and maintenance
We have over 30 years of combined professional research software engineering experience. Our knowledge of the research life cycle allows us to adapt software engineering best practices into workflows to support sustainable software for research.
Please contact us for more information.



