Warm holiday greetings from our team at the Harvard Chan Bioinformatics Core. It’s been a dynamic year at HBC, marked by exciting scientific developments, an evolving team, and fruitful collaborations.
Core. It’s been a dynamic year at HBC, marked by exciting scientific developments, an evolving team, and fruitful collaborations.
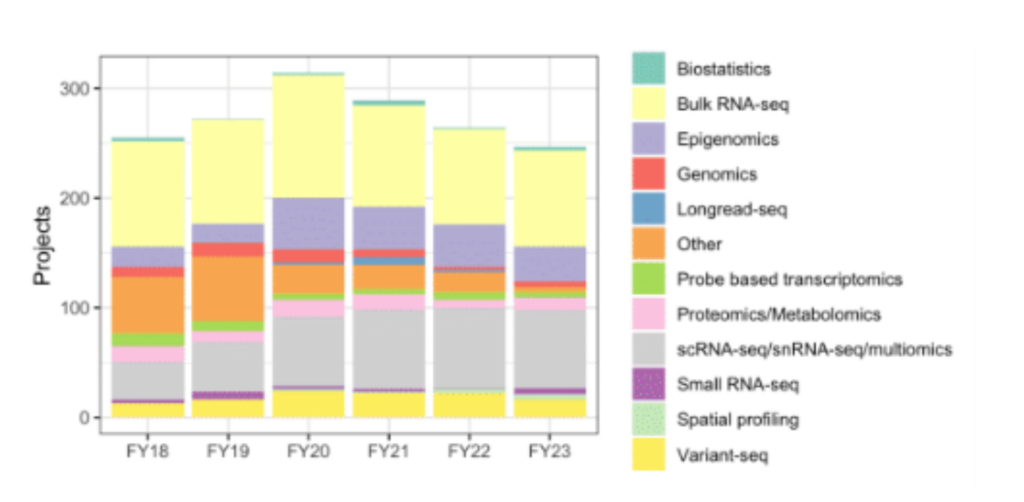
Research: The demand for bulk and single-cell RNAseq analysis, including 10X, CITE-seq, V(D)J, and multimodal analysis, remains strong, enabling our researchers to address key questions about cell type identity, heterogeneity, expression changes, and trajectories. Our core received 270 requests for bioinformatics support this year, including 50 requests for grant support. Core members coauthored 24 publications spanning diverse research areas such as cancer, immunology, evolution, and skin biology. We have also been delving more deeply into spatial transcriptomics, analyzing CosMX, Visium, DBiT-seq and GeoMX experiments, and planning for new studies using the Xenium and Merscope platforms. We are excited to collaborate with the community to explore bioinformatics approaches for spatial transcriptomics and establish robust workflows for analysis.
Infrastructure: Our NGS workflow automation platform, bcbio-nextgen, supported by a CZI “Essential Open Source Software for Science” grant, remains integral to our operations. With 92 references on Google Scholar in 2023, this open-source project continues to be a valuable resource to the international scientific community. Our core team has also been exploring the use of Nextflow for new workflow development after attending the Nextflow Summits in Barcelona and Boston.
Training: Our training team was thrilled to return to in-person workshops this year. We offered 15 workshops serving the greater Harvard community, including four in-person workshops. We continued to teach our full assortment of basic data skills and advanced topics in NGS analysis workshops, while also collaborating on courses with the Center for Computational Biomedicine (CCB), Countway Library Data Services and Department of Biomedical Research (DBMI). We offered 11 half-day modules covering topics in data skills and bioinformatic analyses as part of our popular Current Topics in Bioinformatics series, including a new module on Advanced Shell and a guest presentation by Chris Magnano (CCB) on Machine Learning for Bioinformatics. We were excited to bring back our networking breakfasts to help cultivate a stronger sense of community amongst researchers and Core facilities across campus. We will continue to collaborate and develop new training materials as we encounter data from new platforms, validate novel workflows and establish best practices. Our training materials are freely available and accessible to the community and designed for self-learning.
Personnel: Staffing changes have been a focal point this year. After nine years as our Training Director, Radhika Khetani started an exciting position as Director of Data Science Learning at AstraZeneca, Gothenburg. Meeta Mistry, a co-founder of the training program, brings her exceptional skills and experience to her new role leading our training initiative. Driven by Harvard’s policy shifts for international remote work, we were sad to say goodbye to Amélie Julé who was working from the UK. Victor Barrera (Spain) and Sergey Naumenko (Canada) will depart at year-end. Their deep expertise in transcriptomics, spatial analysis, cancer genomics, and multimodal single-cell RNAseq analysis will be greatly missed. Other notable farewells include Jihe Liu (now a Senior Data Scientist at Cohere Health) and our esteemed Faculty Advisor, Peter Kraft, who joined the National Cancer Institute’s Division of Cancer Epidemiology and Genetics as the Director of the Trans-Divisional Research Program. We are thrilled to have John Quackenbush as our new Faculty Advisor.
This spring, we welcomed Noor Sohail and Heather Wick to our team. Noor comes to us from Dana Pe’er’s group at MSKCC with extensive single cell expertise, and Heather joins us from The Broad, with experience in multiomic data integration. We are actively recruiting and looking for talented, motivated bioinformaticians to join our team.
We were delighted to celebrate Zhu Zhou, Meeta Mistry, and Shannan Ho Sui’s promotions on the Research Scientist track this year, as well as Maria Simoneau’s promotion to Sr. Project Coordinator. Ruben Quinones has joined us as dedicated finance support.
Many thanks and congratulations to our team members for their fantastic work. As we reflect on these changes and advancements, we look forward to the coming year, embracing societal and scientific shifts, from ChatGPT to cutting-edge pipelines. Thank you for your collaborations. We extend warm wishes for a joyful holiday season.



